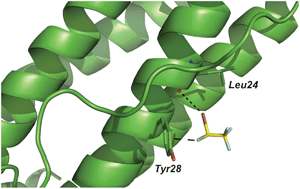
Halogen bonding has been extensively described in the context of a variety of self-assembled supramolecular systems and efficiently utilized in the rational design of materials with specific structural properties. However, it has so far received only little recognition for its possible role in the stabilization of small molecule-protein complexes. In this tutorial review, we provide a few examples of halogen bonds occurring between small halogen-substituted ligands and their biological substrates. Examples were drawn from a diverse set of compounds, ranging from chemical additives and possible environmental agents such as triclosan to pharmacologically active principles such as the volatile anesthetic halothane or HIV-1 reverse transcriptase inhibitors or a subset of non-steroidal anti-inflammatory drugs (NSAIDs) that are halogen-substituted. The crystal structures presented here, where iodine, bromine, or chlorine atoms function as halogen bonding donors and a variety of electron rich sites, such as oxygen, nitrogen and sulfur atoms, as well as aromatic p-electron systems, function as halogen bonding acceptors, prove how halogen bonds can occur in biological systems and provide a class of highly directional stabilizing contacts that can be exploited in the process of rational drug design.